Morphology is the study of shape.Mathematical morphology is a process where we take in binary images and operate on them to improve the image before further processing or to extract information from the image. The process' language is the set theory such that morphological operators take a binary image and a structuring element (or kernel but the term is mostly used in convolution). A point is either in the pixel set such that it is part of the foreground or not such the pixel is part of the background. Because of this, set operations such as intersection, union, inclusion and complement are applied. For more information, one can look up Mathematical Morphology.
The structuring element consists only of 1's and don't care pixels or pixels we can ignore. It is shifted across the image and at every pixel of the image it is compared with the ones underneath.If the condition defined by the set operator are met(e.g. the set of pixels in the structuring element is a subset of the underlying image pixels), the pixel underneath the origin of the structuring element is set to a pre-defined value (0 or 1 for binary images).[2]
The desired resulting image is therefore heavily dependent on the structuring element used.
In this activity, we look at morphological operations on the following shapes in Figure 1 with structuring elements which followed. In the first part, we were asked to draw the results to maybe check if we got the concept right. Later on, Scilab can help us check if we did.
 |
| Figure 1. Shapes |
 |
| Figure 2. Structure Elements |
Erosion denoted by an enclosed minus sign erodes the boundary of the foreground pixels which makes the area of the foreground smaller and the spaces in between become larger. Dilation denoted by an enclosed plus sign on the other hand, does the opposite which expands the foreground and diminishes the spaces in between.
Below are my drawings and I opted to place them separately to maintain the size for easy viewing.
I. Erosion
The eroded part is indicated by the boxes with "-". In each structuring element, the origin is indicated by a dot since we are dealing with operations that are based on the set theory which is dependent on a reference point.
 |
| Figure 3. Erosion of 5x5 Square |
 |
| Figure 4. Erosion of Triangle |
 |
| Figure 5. Erosion of 10x10 Hollow Square |
 |
| Figure 6. Erosion of Plus Sign |
II. Dilation
Dilated parts are indicated by the "+"
Dilated parts are indicated by the "+"
 |
| Figure 7. Dilation of Square and Triangle |
 |
| Figure 8. Dilation of 10x10 Hollow Square |
 |
| Figure 9. Dilation of Plus Sign |
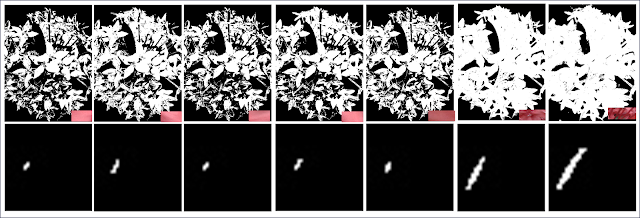
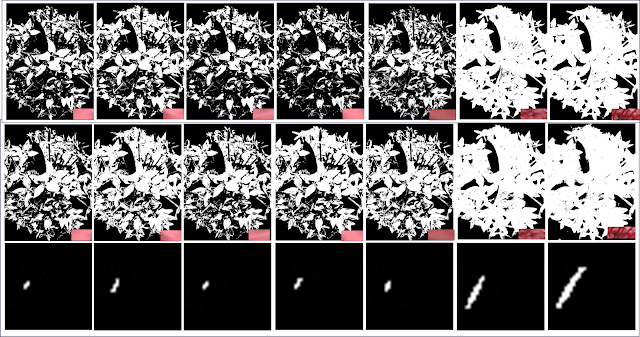
To see if I predicted the output correctly, erosion and dilation were performed in Scilab. Figure 10a-10b are the results for the erosion 5x5 square with the structuring elements in the following order: 2x2 ones, 1x2 ones, 2x1 ones, cross, and diagonal which is 2 boxes-long. The same is done with the others. Figure 10e-10i Erosion of a B=4, H=3 Triangle; Figure 10j-10n Erosion of 10x10 hollow square; and Figure 10o-10s. Erosion of plus sign. Figure 11 displays the results for dilation and obeys the same ordering.
 |
| Figure 10. Erosion of the shapes with the structure elements performed in Scilab |
 |
| Figure 11. Dilation of the shapes with the structure elements performed in Scilab |
Now that we know how erosion and dilation works on images, we look at the different morphological operators: Closing, Opening, Tophat (White and Black). There are instances that there is an overlap in the graylevel distribution of background that sometimes result to artifacts after segmentation and binarization. The aformentioned operators will help us solve this problem.
 |
| Figure 12. Circles002.jpg |
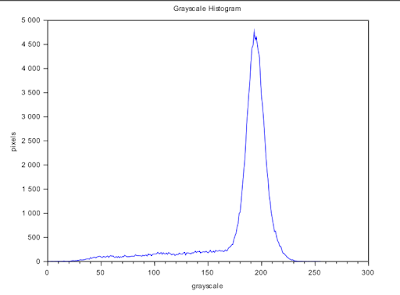
To determine how these operators work, we slice the image in Figure 12 into 256x256 subimages. We take the first subimage which is the first 256x256 slice at the topleft corner of Circles002.jpg. To separate the foreground from the background, we use the method of thresholding aided by a graylevel histogram as shown in Figure 13. We see that the appropriate threshold value is ~210 or 0.8235 if we use the im2bw() function.
 |
| Figure 13. Graylevel histogram of a subimage |
As said earlier, binarizing an image may result to artifacts as seen in Figure 14: white areas on the upper edge and white dots can be seen on areas where we expect it to be black or zero. Since we are only interested with the white circles, we want to remove these artifacts.
 |
| Figure 14. 256x256 subimage considered from the top left portion of the iamge |
 |
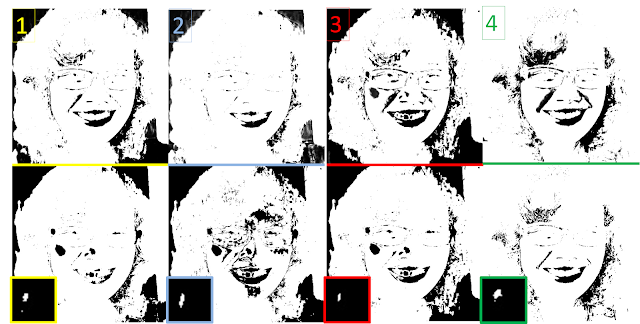
| Figure 15. a) Subimage operated with different morphological operators:(b) Closing (c) Opening (d) Black Tophat (e) White Tophat |
Below is a simple description of what the respective operators do:
1. Closing operator = CloseImage(subbw,struc) = erode(dilate(subbw,struc))
2. Opening operator = OpenImage(subbw,struc) = dilate(erode(subbw,struc))
3. White Tophat operator = subbw - OpenImage(subbw,struc)
4. Black Tophat operator = CloseImage(subbw, struc) - subbw
As we can see the Opening operator was successful in removing the artifacts and isolating the circles. In the IPD package of Scilab, one may use the function SearchBlobs() to label each aggregate of 1's in an image. This function aids us determine the area of each aggregate where the area refers to the sum of all ones.
 |
| Figure 16. Searchblobs function labels each blob of white pixels for easier analysis |
Using the function, we take the area values of all the blobs in the subimages by scanning through all the aggregates from i = 1: max(Searchblobs(bw)) and plot its histogram.
 |
| Figure 17. Histogram of areas in the subimages |
From this distribution, we can get the mean and standard value of the areas in the images. It was found that the mean $\mu = 643.15$ and the standard deviation $\sigma = 455.39$. From these values, we can determine circles which are abnormal by filtering the areas above the sum of the mean and standard deviation.
Now we know about structuring elements and what the operators do. We also know the properties of the SearchBlobs() function. It's time to apply what we know to something practical with very much importance--the identification of cancer cells.
Figure 18 shows white circles with several larger circles which we identify as cancer cells. We want to filter these cancer cells using the techniques we learned earlier in the activity.
 |
| Figure 18. Circles with cancer cells.jpg |
Below shows the binarized image and the identified blobs with areas exceeding the Threshold = mean(area) + stdev(area) with the use of the function in IPD, FilterBySize(bwlabel,Threshold) where bwlabel = SearchBlobs(bw). The function removes all the cells whose areas are below the threshold. The ones whose areas are above it are deemed abnormal. But as we shall see, because of the opening operator which erodes and then dilates causing the circles to get connected, aggregates are formed by normal cells which are retained by the FilterBySize() function. The next thing we need to do then is to separate these normal cells. Maam Jing suggested the use of Watershed Segmentation technique which you can read and learn using this tutorial: IPD Tutorial.
 |
| Figure 20. a)Binarized image b) Identified cells with area larger than the sum of mean and standard deviation c) Same with that in (b) but now we manipulate the graylevel value to see them easily |
In class, I was doing the watershed segmentation part but luckily, because Maam Jing is doing her rounds, I was able to consult her about something simple that I did which automatically isolated the cells! I was very skeptic at first because the motivation that's in my head during the activity is to learn how to automate. Things like cancer are serious and needs fast action. Human error is not forgivable when lives are at stake. (Huuu I remember my med dreams but no.. I'm fineeee :)).
Well, back to the simple something. Remember, in the earlier part, I said something about the structuring element being important to get the desired result? Well, I proved its importance very quickly by using a circle as my structuring element whose radius is $r=15$. And tadddaaaa! I was able to get all five of them unlike the prior method that I used which only retained 3 out of 5! I was of course, pretty skeptic at first, but according to Maam Jing,the method which requires the least user input is already fine. So there, I have it. Yey! The identification of the cancer cells was a success!
Well, back to the simple something. Remember, in the earlier part, I said something about the structuring element being important to get the desired result? Well, I proved its importance very quickly by using a circle as my structuring element whose radius is $r=15$. And tadddaaaa! I was able to get all five of them unlike the prior method that I used which only retained 3 out of 5! I was of course, pretty skeptic at first, but according to Maam Jing,the method which requires the least user input is already fine. So there, I have it. Yey! The identification of the cancer cells was a success!
 |
| Figure 19. Image with the identified cancer cells |
Now that I'm done with my activity, I can now go and explore. :D One quick thing that came to my mind while I was doing the activity is to apply morphological operations on my research. My research is on cities and I take interest in roads which are a city's one of, if not the most long lasting element. In particular, I want to apply the morphological operations to obtain a road network which is purely composed of one-pixel-thick lines. This is because, as we know, roads may contain islands. In my shapefiles, these islands will be rendered as a space in between the lines or, roads maybe multi-lane so that its thickness contributes much to space-filling. One of the metrics that I'm using to characterize my cities is the fractal dimension, $f_D$. My goal is to count the white pixels that cover the entire length of roads and not taking into account the thickness so much because the thickness will inflate the number of pixels and may return a very big value for the $f_D$. This will probably give a wrong impression of over space-filling!
So there, considering a map image of Quezon City as seen in Figure 20. What I want to get is a road network that is purely composed of 1-pixel thick lines.
 |
| Figure 20. Map of Quezon City |
 |
| Figure 21. Binary image of QC map |
Zooming into Figure 21,we will see roads which has spaces in between them.
Now, I apply the closing operator, which as you may recall is equivalent to Closing = ErodeImage(DilateImage(im,struc)) where the structuring element that I used is a 1x2 ones. I got the following result:
 |
| Figure 22. Resulting image using the closing operator with a 1x2 ones structuring element |
The difference is not very obvious, but zooming in, we will see that the thin spaces in between have closed and that the road network is now composed of approximately purely lines.
I also found a morphological operator in Matlab, 'skel' which is a part of the bwmorph() function which maybe the more appropriate operator for my problem. 'Skel' removes the pixels on the boundaries of objects but does not allow them to break apart. The remaining make up the image skeleton.
References:
[1] M. Soriano, "Morphological Operations",AP 186 class manual.(2016)
[2] R. Fisher,S. Perkins,(2003) "Morphology", retrieved from http://homepages.inf.ed.ac.uk/rbf/HIPR2/morops.htm on 22 October 2016.
[3] P.Peterlin,"Morphological Operations: An Overview", retrieved from http://www.inf.u- szeged.hu/ssip/1996/morpho/morphology.html on 22 October 2016